The last few days I’ve spent some time looking at how parallelism in yt performs. I conducted two different tests, both of which operated on the 512^3, 7 level ‘Santa Fe Light Cone’ dataset RD0036. This dataset has 5.5e8 total cells and in the neighborhood of 380,000 grids. I ran four different tests: a 1D profile of the entire dataset, a 2D profile of the entire dataset, and projections of both ‘Density’ (requires IO) and ‘Ones’ (doesn’t require IO). For the purposes of these tests I have excised any time spent in creating the hierarchy and affiliated grid objects. The results are shown just here:
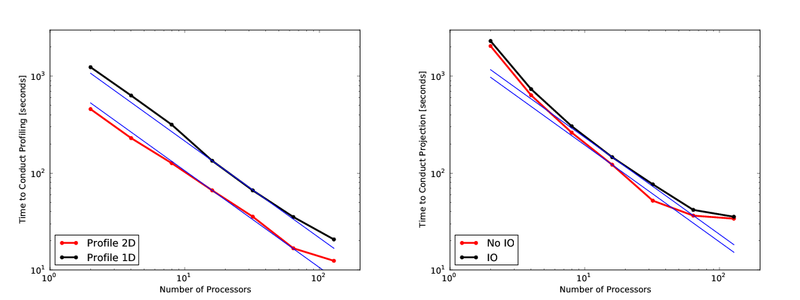
One thing I found rather astonishing about this is how poorly the 1D profile operates. It’s not incomprehensibly slow, but it operates much slower than the 2D profile. I investigated why, and I believe that it comes down to a fundamental difference in the implementation. The 2D profile’s binning algorithm is implemented in C, whereas the binning algorithm in the 1D profile is actually implemented in Python, such that it actually scales quite poorly with the number of bins. Since I was taking 128 bin profile, this enhanced the problem substantially. Regardless, while a new 1D profiling implementation probably won’t be written for yt-2.0 (as it’s not so slow as to be prohibitively costly) it will be replaced in the future.
I was, however, quite pleased to see the parallelism scale so nicely for the profiles. Projections had a bit of a different story, in that their strong scaling saturated at around 64 processors; I believe this is probably due simply to the projection algorithm’s cost/benefit ratio. I would point out that in my tests there was some noise in the high-end – in one of the runs, projecting on 128 processors took a mere 6 seconds. Then again, since you only need to project once anyway (and then spend quite a while writing a paper!) the time difference between 30 seconds and 6 seconds is pretty much a wash.
Now, before I get too enthusiastic about this, I did find a couple very disturbing problems with the parallelism in yt. Both were within the derived quantities – these are quantities that hang off an object, like sphere.quantities[‘Extrema’](‘Density’) and so on. (In fact, that’s the one that brought the problem to light.) I was performing a call to ‘Extrema’ before conducting the profiling step and I saw that it took orders of magnitude more time than the profiling itself! But why should this be, when they both touch the disk roughly the same amount? So I dug in a bit further. Derived quantities are calculated in two steps. The first one is to calculate reduced products on every grid object: this would be the min/max for a given grid, for the above example of ‘Extrema’. All of the intermediate results then get fed into a single ‘combine’ operation, which calculates the final result. This enables better parallelism as well as grid-by-grid calculation – much easier on the memory!
Now, in parallel we usually preload all the data that is going to be accessed by a given task. For instance, if we’re calculating the extrema of density, each MPI task will read from disk all the densities in the grids it’s going to encounter. (In serial this is much less wise, and so we avoid it – although batch-preloading could be a useful enhancement in the future.) However, what I discovered was that in fact the entire process of preloading was disabled by default. So instead of conducting maybe 10-30 IO operations, each processor was conducting more like 10,000. This was why it was so slow! I’ve since changed the default for parallel runs to be to indeed preload data.
The other issue was with the communication. Derived quantities were still using a very old, primitive version of parallelism that I implemented more than two years ago. In this primitive version, the root processor would turn-by-turn communicate with each of the other processors, and then finally broadcast a result back to all of them. I have since changed this to be an Allgather operation, which enables collective broadcasting. This sped things up by a factor of 4, although I am not sure that in the long run this will be wise at high processor counts. (As a side note, in converting the derived quantities to Allgather, I also converted projections to use Allgatherv instead of Alltoallv. This should dramatically improve communication in projections, but communication was never the weak spot in projections anyway.)
There are still many places parallelism could be improved, but with just these two changes to the derived quantity parallelism, I see speeds that are much more in line with the amount of IO and processing that occurs.